May 2024
Project 8p Research Newsletter
💬 Research Question of the Month
There’s a perennial thorny question we keep returning to over and over again in our sponsored research projects: how similar are 8p heroes to each other? Initially, all of our studies focused on a single pioneer invdupdel 8p family trio — mom, dad, and affected child — for practical reasons. Over time, Project 8p’s biobank has grown to include additional 8p hero cell lines, so we now have a more representative census of the 8p hero population. Highlighted below are results from Professor Aryeh Warmflash’s lab at Rice University that start to chip away at this question about similarities (and differences) between 8p heroes.
Please drop feedback in the comments!
🔬Research Highlight
Project 8p funded Professor Warmflash’s lab to study early fetal development in the lab using induced pluripotent stem cells (iPSCs) derived from 8p heroes. The Warmflash lab specializes in a technique called the germ layer micropatterning assay, which offers a window into how the growing embryo organizes itself.
A quick refresher on developmental biology. All organ systems of the body originate from one of the three germ layers: ectoderm (outer layer), mesoderm (middle layer), and endoderm (inner layer). The endoderm gives rise to digestive and respiratory systems. The mesoderm gives rise to the skeleton, muscle, heart and bones. The ectoderm births the nervous system and the skin.
Prof Warmflash previously presented data at one of our monthly Research Roundtables on WES557, the pioneer invdupdel 8p hero that kickstarted all of our research projects. Project 8p sponsored this project in order to shed light on the earliest changes that occur in human development as a result of complex 8p rearrangements. Although this research won’t immediately translate to therapies, it’s the foundation on top of which medicines might one day be made.
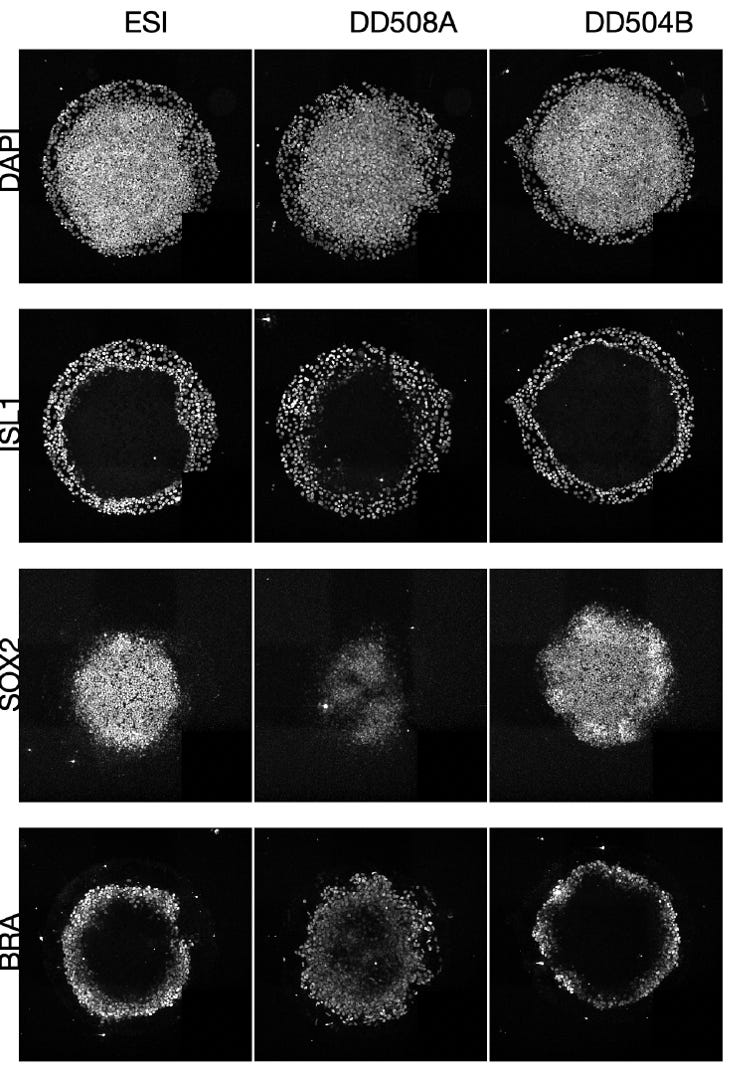
As shown in the summary figure below, the WES557 has three distinct phenotypes that are easily discerned by eye: overgrowth of the mesoderm layer (blue), expansion of the extra-embryonic layer (red), and contraction of pluripotent cells (green).
Are these phenotypes affecting the 3-D organization and size of the germ layers unique to invdupdel 8p heroes? What would another invdupdel 8p hero sample look like? And what about deletion-only 8p heroes? Or duplication-only 8p heroes?
Below are results of a micropatterning assay that compared an unaffected control (ESI) to a second invdupdel 8p hero line (DD508A) and a deletion-only 8p hero line (DD504B). DD508A looks strikingly similar to WES557. We obviously need to process more invdupdel samples to make a firm conclusion but we can say a trend is emerging. Fascinatingly, the deletion-only 8p hero line DD504B does not look like a invdupdel 8p hero sample. In fact, DD504B appears more similar to the unaffected control.
TNKS1 is a candidate 8p driver gene that we’ve had on a radar. It’s typically in the deleted 8p segment resulting in haploinsufficiency, i.e., having just one copy of the gene versus the normal complement of two copies. The Warmflash lab sought to isolate the effects of TNKS1 haploinsufficiency (TNKS+/-) and compare them to the effects of knocking out both copies of TNKS1 (TNKS-/-). These experiments were not performed in 8p cell lines. Rather, one or both copies of TNKS1 were purposefully deleted in an unaffected wildtype control (WT) line. The results below show that TNKS1 haploinsufficiency (TNKS+/-) causes a reduction of the mesoderm phenotype, i.e., lower BRA signal intensity, which is further intensified in the TNKS-/- knockout.
The deletion-only 8p hero line DD504B shows a reduced ring of primitive streak mesoderm marked by BRA, to use an embryologist’s parlance. This result is consistent with the data on the TNKS1 knockout, which shows a similar phenotype. Therefore, deletion of TNKS1 may partially explain the DD504B germ-layer phenotype.
📄 Recent Articles and Publications
Let us know if you’ve read or published a paper or article recently that you think the 8p community should know about!
Earlier this month, we published a Substack about the manhunt for 8p driver genes. We introduced the concept of driver genes that are not physically located on the short arm of chromosome 8 aka backseat drivers.
Chromosome Transplantation: Opportunities and Limitations. La Grua et al. Cells. April 2024. https://www.mdpi.com/2073-4409/13/8/666
Nice review article that summarizes the state of play in what we’ve always referred to as chromosome therapy.An asymptomatic male individual carrying a 5.72 Mb de novo deletion in 8p23.2‑p23.3: A case report. Keramida et al. Experimental and Therapeutic Medicine. June 2024. https://www.spandidos-publications.com/10.3892/etm.2024.12529
Interesting case report of a 30-year-old asymptomatic male with a 5.721 Mb deletion in the 8p23.2‑p23.3 region, including ARHGEF10, CLN8, CSMD1, DLGAP2, ERICH1, FBXO25, KBTBD11, MYOM2, TDRP, and ZNF596. The normal phenotype of the patient supports the hypothesis that there is incomplete penetrance of 8p23.2‑p23.3 deletions, which means there may be “anti-drivers” out there: genes that prevent or blunt the effects of genomic imbalance.
💜 Family Corner
Last week, the Project 8P team hosted our Q2 Family Update, unveiling the expansion of our natural history study. Together, we're venturing into exciting new opportunities! Missed the update? Don't worry! Email kaiti@project8p.org to catch up on all the details.
📆 Upcoming Events
Project 8p Research Roundtable, May 30th, at 12pm EST on Zoom. Email barbara@project8p.org to attend.