Skin deep
Disease modeling in 8p brain organoids led to the discovery of Reelin as a novel potential driver gene. Will skin cells from 8p heroes teach us about the disease and lead to treatments? We think so.
In collaboration with
Project 8p is the world’s first nonprofit biotech whose mission is to lead the charge toward treatments and cures for complex chromosome diseases caused by megabase rearrangements. Pick your flavor combo: deletion, duplication, and even inversion. 8p heroes can have all three, including heroes who just have an 8p deletion or an 8p duplication.
For some perspective on scale, and as another reminder why 8p is particularly challenging: you can fit an entire copy of human chromosome 21, which encodes several hundred genes, inside a single typical 8p duplicated segment.

A year ago, Project 8p commissioned the first edition of a CureMap, which identified five interlocking research priorities that assemble into a superstructure, much like the segments of a chromosome. Disease modeling is the centromere, the core scaffold around which the rest of the chromosome is constructed.
Project 8p placed a bet on Dr Alysson Muotri’s lab at UCSD to conduct a first-of-its-kind neurodevelopmental disease modeling study on a pioneer 8p family trio. Specifically, induced pluripotent stem cells (iPSCs) from mom, dad and affected child were differentiated into 1-month, 3-month, 6-month, and 9-month-old brain organoids. Brain organoids were disaggregated and then single-cell RNA-seq was performed.
The bet paid off with the surprising discovery of Reelin (RELN) over-expression in a large population of differentiated glutamatergic neurons in the proband’s 9-month 8p brain organoids that appear to be Cajal-Retzius cells. There’s also a second but smaller population of Reelin-positive glutamatergic neurons in the proband’s 9-month 8p brain organoids that are actively dividing. Followup electrophysiology studies are already underway in Dr Muotri’s lab. Project 8p is also working with Dr Hiruy Meharena’s lab at UCSD, which has been studying aneuploidy from the trisomy 21 perspective.
In this update, we’ll highlight a complementary approach to 8p disease modeling using 8p hero-derived skin cells aka fibroblasts. At first blush, fibroblasts may pale in comparison to brain organoids but fibroblasts have a few tricks up their sleeve. Like enabling drug repurposing screens faster and cheaper than any other 8p disease model available. And serving as a launchpad to studies in slower and costlier 8p disease models.
The technical term for complex chromosome rearrangements is aneuploidy. The legendary trailblazing aneuploidy scientist and yeast geneticist Dr Angelika Amon, whom the world tragically lost to cancer in October 2020, once defined aneuploidy as:
A change in chromosome number that is not the exact multiple of the haploid karyotype is known as aneuploidy. This condition interferes with growth and development of an organism and is a common characteristic of solid tumors.
Because of her expertise and reputation, one of the first people contacted by Project 8p’s founder was Dr Amon, whose lab was located at the Massachusetts Institute of Technology. Years ago, using the budding yeast Saccharomyces cerevisiae as a model system, Dr Amon’s lab published numerous studies showing that proteotoxic stress, increased glucose uptake and other changes in basic cell physiology occur when a chromosome is out of balance. (Her lab was the first to identify suppressor mutations that allow cells to tolerate aneuploidy!)
However, a general theory of aneuploidy that explains the vulnerabilities of specific cells to specific chromosome imbalances at specific times throughout the life of an organism has so far eluded scientists. There are so many gears and mechanisms in constant motion. And yet there’s hope.
Despite having unique chromosomal breakpoints and non—overlapping sets of affected genes, 8p heroes share a constellation of symptoms. Is Nature telling us that if you break the short arm of the 8th chromosome in any manner, the outputs converge into 8p disease?
Even if there are indeed individual culpable driver genes on 8p — or lurking on other chromosomes — that are responsible for one of more aspects of disease, does the collective action of these drivers explain everything, or is chromosome imbalance itself a primary disease driver? If the answer tends toward the latter, then the implication is that complex chromosome diseases may be united by one or more common mechanisms of disease onset and progression.
Before the brain organoid bet, the first 8p disease models were actually a pioneer 8p family trio of skin fibroblasts studied in the Amon lab at MIT in 2019 and 2020. Specifically, bulk RNA-seq was performed on said 8p trio in order to assess how the expression of each gene in the genome is altered in: i) 8p fibroblasts; ii) unaffected age-matched controls; iii) fibroblasts derived from people living with trisomy 21 aka Down Syndrome.
The following analysis was performed by Perlara Cure Guide Dr Arun Ramani. In keeping with Project 8p’s open science credo, these data will soon be available for download on the 8p online data portal.
Hierarchical clustering shows that three different 8p fibroblast lines and three different T21 fibroblast lines form sister clades. Three out of four wildtype fibroblast lines co-cluster and form an out-group relative to 8p and T21. One of the four wildtype fibroblast lines wedged itself into the T21 cluster.
Another way to flatten high-dimensional dataset into a meaningful 2-D figure is principal component analysis (PCA). Mirroring the results of hierarchical clustering, 8p and wildtype fibroblasts form distinct groups separated in PCA space. The three 8p fibroblast lines are huddled closely together. The three wildtype fibroblast lines are a bit more spaced out. The T21 fibroblast lines are stretched out along both PCA1 and PCA2 axes, but overall more similar to the 8p lines than to the wildtype lines.
So how similar are 8p fibroblasts and T21 fibroblasts to each other? We generated lists of differentially expressed genes and visualized the results as Venn diagrams. Let’s start with genes whose expression goes down. There are 161 genes that are down-regulated in both 8p and T21 relative to wildtype, which translates to 60% of all 8p-down-regulated genes and 40% of all T21-down-regulated genes. That quite a bit of overlap between 8p and T21!
Now let’s examine the genes whose expression goes up. The association gets even stronger. There are 404 genes that are up-regulated in both 8p and T21 relative to wildtype, which translates to 69% of all 8p-down-regulated genes and 52% of all T21-down-regulated genes.
Shifting gears to focus on the comparison between 8p and wildtype, a volcano plot was generated comparing gene expression of wildtype fibroblasts versus 8p fibroblasts. Some of the differentially expressed genes are labeled. There are hundreds of differentially expressed genes above the p-value and 2-fold change thresholds, so we turned to pathway analysis to glean insights — too many to process individually.
Gene set enrichment shows that known aneuploidy-sensitive cell physiological processes like protein translation, organelle biogenesis and mRNA splicing are affected in 8p. Following up on the organelle biogenesis and maintenance gene ontology, mitochondrial genes are strongly down-regulated in 8p fibroblasts relative to wildtype fibroblasts.
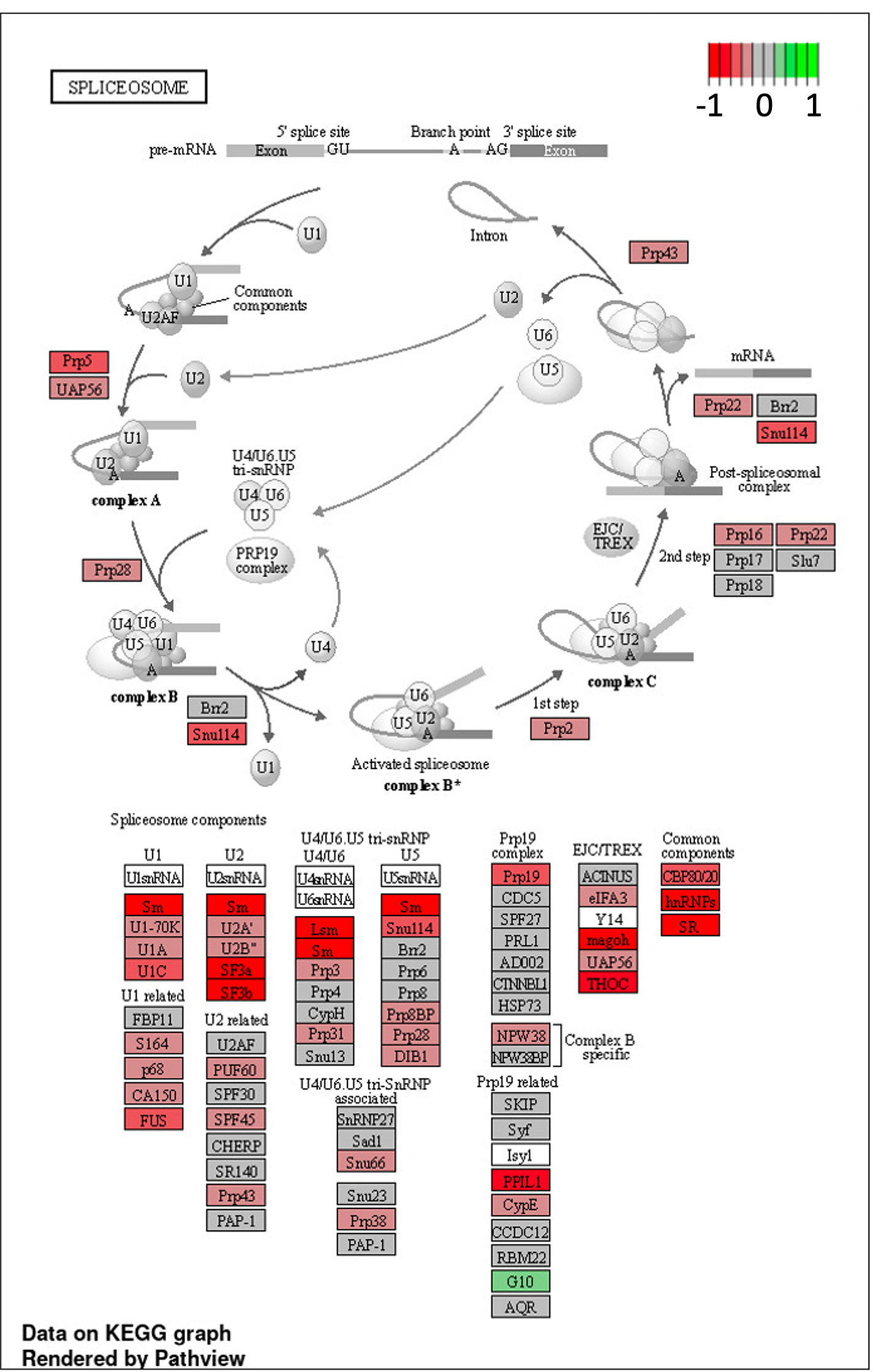
We look closer at a single cellular component called the spliceosome, which as its name suggests in the site of action for processing mRNA into its final form. The building blocks of spliceosomes are strongly down-regulated in 8p fibroblasts relative to wildtype fibroblasts.
While we’re still some ways from a general theory of aneuploidy, it’s clear from studying gene expression changes in skin cells from 8p heroes that chromosome imbalance itself can be considered a primary disease driver. Keep reading to find out what we do with this insight.
It’s not practical to use RNA-seq as a readout for a high-throughput drug repurposing screen. Based on precedents with skin fibroblasts in perhaps the most well-studied complex chromosome disease — trisomy 21 (T21) aka Down Syndrome — Project 8p sponsored a cell biological workup on 8p fibroblasts in Dr Nicoleta Moisoi’s lab at De Montfort University in the UK.
Dr. Marta Dominguez-Prieto in the Moisoi lab put 8p hero skin fibroblasts through a series of stress tests alongside age-matched control fibroblasts from healthy donors. Fibroblast lines from four 8p heroes were compared to fibroblast lines from four age-matched controls, as well as the two parental lines that form a trio with one of the 8p hero lines.

Both control and 8p fibroblasts were subjected to a battery of image-based cellular assays, including the Cell Painting panel of vital dyes that was originally developed in Dr Anne Carpenter’s lab at the Broad Institute. Mitochondrial function in particular was examined given that aneuploidy is known to affect cellular metabolism. Growth in media supplemented with glucose was also compared to growth in media supplemented with galactose, which forces cells to use their mitochondria for energy production.
Mitochondrial resting membrane potential was assessed using the fluorescent probes TMRM and TMRE. Results for TMRE are shown below (and the same result was observed using TMRM). In both glucose and galactose conditions, 8p fibroblasts have lower mitochondrial resting membrane potential than control fibroblasts. These results are reassuringly consistent with Arun’s analysis of the 8p fibroblast RNA-seq dataset.
Next, each of the Cell Painting dyes was evaluated in either glucose or galactose media. 8p cells exhibited a phenotype, i.e., looked different than control fibroblasts, more robustly and reproducibly in galactose media than in glucose media. Sometimes the 8p lines showed a reduction in signal intensity, other times the 8p lines showed an increase in signal intensity. In the series of figures below, a stock image of a generic cell stained with the corresponding dye or probe is displayed for reference.
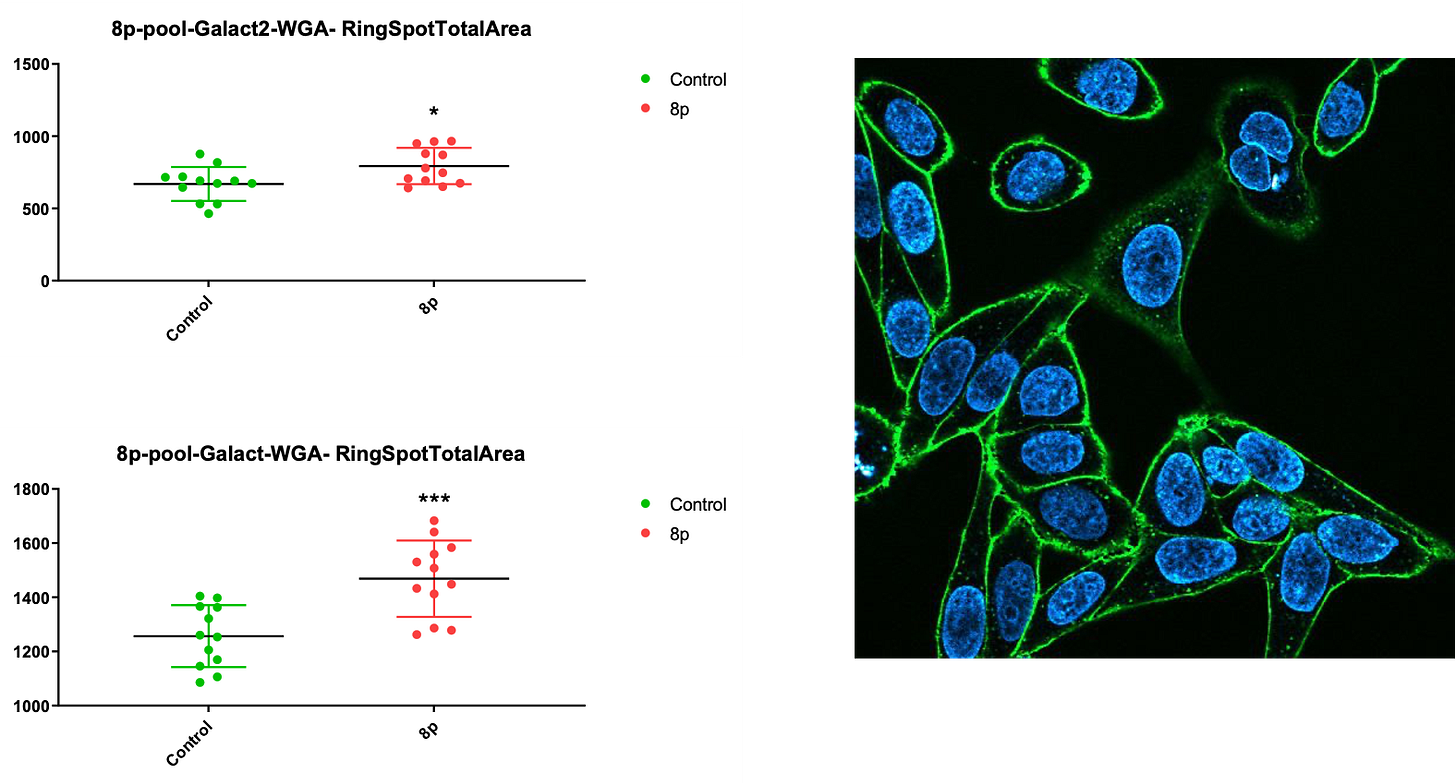
For example, wheat germ agglutinin (WGA), which stains glycoproteins and glycolipids in the plasma membrane, stains 8p fibroblasts more intensely than control fibroblasts only in galactose media in two independent experiments.
Using Lysotracker to stain lysosomes, we observed the same pattern of effect only in galactose media. Increased Lysotracker staining is indicative of autophagic flux and increased lysosomal degradation.
Conversely, concanavalin A, which primarily stains glycoproteins and glycolipids in the endoplasmic reticulum, has the opposite effect. It increases signal intensity in 8p fibroblasts grown in glucose media but not in galactose media.
All in all, these exploratory assay optimization experiments give us confidence that Cell Painting features will distinguish 8p fibroblasts vs control fibroblasts. However, no single readout above resulted in complete statistical separation of 8p fibroblasts vs control fibroblasts, justifying the use of the Cell Painting panel instead of relying on rescue of a single phenotype.
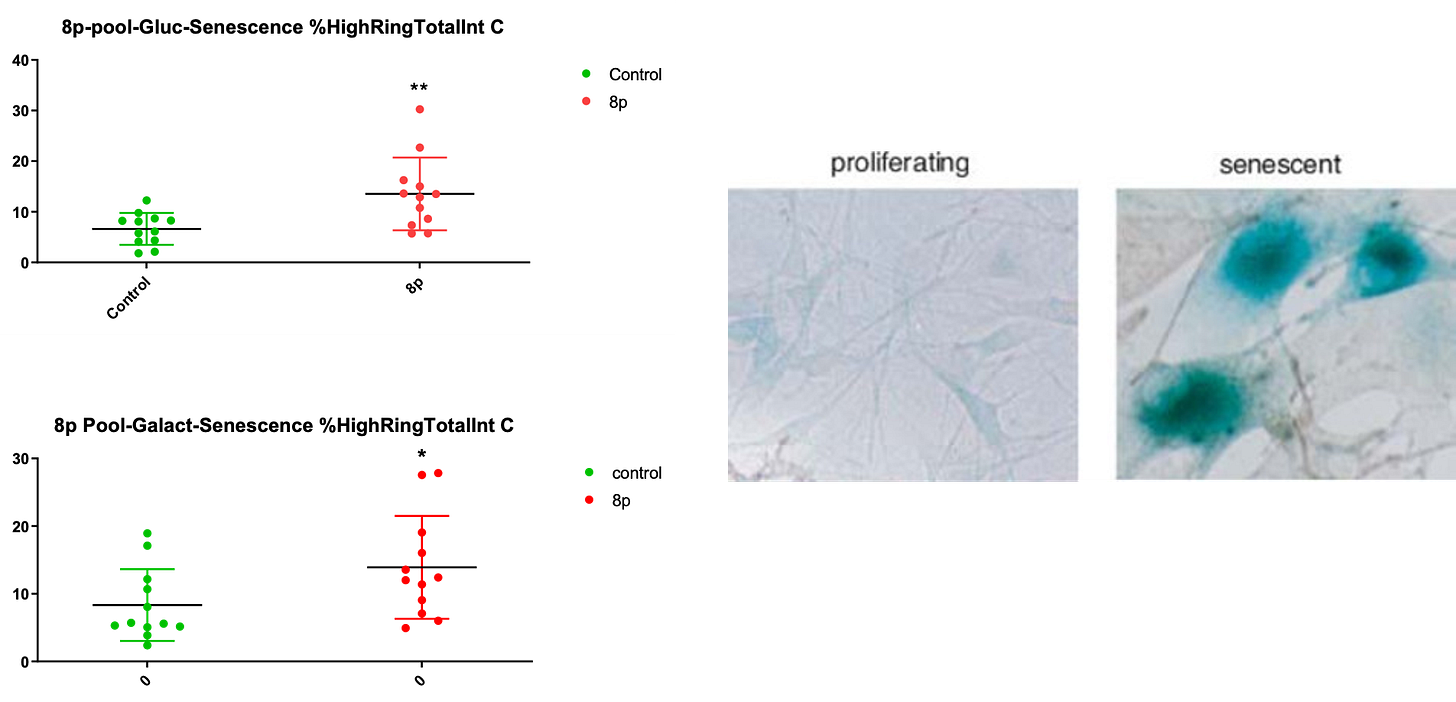
Finally, we assessed whether 8p fibroblasts exhibited higher rates of senescence because there’s a vast literature on the relationship between aneuploidy and senescence. We observed a modest increase in the fraction of senescent 8p cells grown in either glucose media or galactose media.
Stay tuned for a refresh of the 8p CureMap as we approach the midway point of the inaugural two-year research cycle.