A Grand Unified Theory of Neurodevelopmental Chromosomal Disorders
Project 8p's quest is to mend broken chromosomes. By making medicines for 8p heroes living with the fallout of genomic imbalance we might just solve one of biology's most complex puzzles.
The p in 8p is short for petit but it could easily stand for puzzle.
In analogy to Newtonian mechanics failing to predict the composition and motion of subatomic particles, the classical frameworks of aneuploidy fail to pinpoint a minimal set of common driver genes that are responsible for the shared constellation of symptoms and disabilities experienced by all 8p heroes who suffered a pre-conception traumatic genome injury to the short arm of the maternal copy of chromosome 8.
Deletions, duplications and inversions — collectively referred to as copy number variants (CNVs) or complex chromosomal rearrangements — wreak havoc on a scale of one to forty megabases, embroiling hundreds of potential drivers on chromosome 8p alone, not to mention possible drivers lurking on other chromosomes.
Drivers, broadly defined, could be coding genes that make a protein, gene regulatory elements like promoters and enhancers, so-called noncoding genes that don’t encode protein, or other functional sequences.
Over half of 8p heroes suffer all three types of traumatic genome injury involving an enormous span of DNA equivalent to the entire 21st chromosome, which causes Down Syndrome when triplicated.
The typical 8p hero genotype is distal deletion at the tip of the short arm paired with an inverted duplication that variably extends down the short arm of chromosome 8 toward the centromere.
For the aficionados in the audience, the deletions tend to occur during maternal meiosis I between non-sister chromatids of homologous pairs. Duplications tend to occur because the repetitive defensin gene cluster creates fault lines along chromosome 8 where slippages and breakages are more likely to happen.
Unlike other chromosomal disorders that involve smaller CNVs or simpler rearrangements, visual inspection of each 8p hero’s chromosomal breakpoints reveals no single affected stretch of the short arm of chromosome 8 — also known as a critical region — shared by all 8p heroes.
Therein lies the puzzle. At no point along the short arm of chromosome 8 can a straight line intersect the affected segments of all 8p heroes.
In 2021, the first clinical cohort of ninety-seven (97) 8p heroes was described in a study recruited and funded by Project 8p — hereafter referred to as Okur et al — led by geneticist Dr. Wendy Chung, Chair of Pediatrics at Boston Children's Hospital.
A complex clinical presentation includes but is not limited to global developmental delays, intellectual disability, autism spectrum behavior, hypotonia, GI dysfunction, congenital heart defects and seizures.
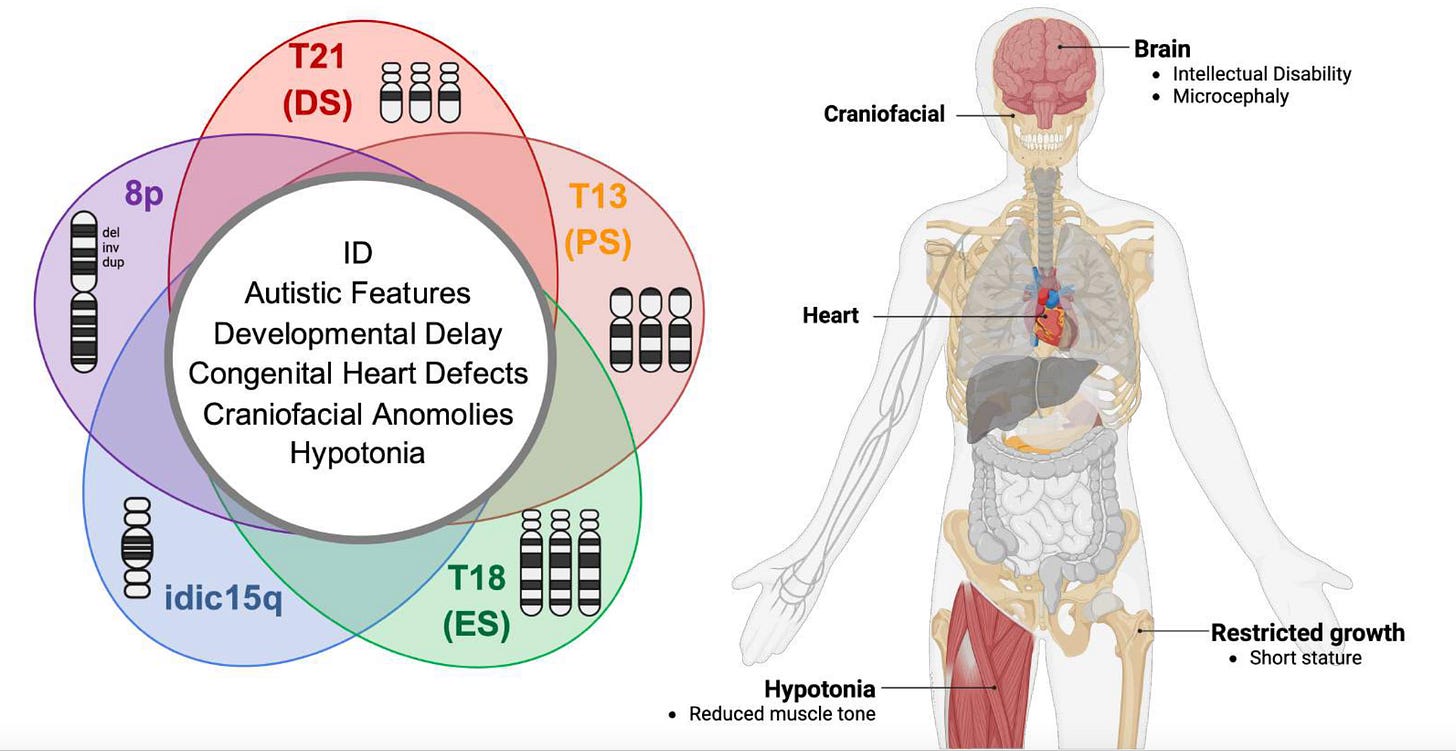
Notably, those clinical features are shared with four other chromosomal disorders: trisomy 21 (Down Syndrome), trisomy 13, (Patau Syndrome), trisomy 18 (Edwards Syndrome), and idic15q (Dup15q Syndrome).
Okur et al identified four critical regions on 8p:
a deleted segment and an inverted duplicated segment shared by invdupdel 8p heroes
a distal deleted segment shared by some del only 8p heroes
a proximal deleted segment shared by some del only 8p heroes
an extended deleted segment shared by some del only 8p heroes
The authors noted the puzzling absence of top dog driver genes in the discussion section of their paper. The genomic equivalent of spooky action at a distance:
This variability of the distal breakpoints further confounds the identification of additional candidate genes beyond shared duplicated segment given that the same major clinical findings are reported in individuals with the smallest and largest duplications although the severity differs with the distal duplication breakpoints. Thus, in addition to the gene(s) within the shared duplicated segment in invdupdel(8p), involvement of additional genes beyond the shared duplicated segment such as CHRNA2 may add incrementally to the clinical spectrum.
Since 2022, Project 8p sponsorship of research has resulted in a foundational dataset for a pathfinding invdupdel family trio of biosamples obtained from 8p hero, mother and father.
We now have the biobank and operational playbook to scale up experiments that were prototyped on the inaugural 8p family trio. Over the course of 2024, Project 8p will fund new research that will assess the cellular and clinical convergences not only between 8p heroes, but also between 8p and other chromosomal disorders.
In the history of science, solving a difficult puzzle often births a paradigm shift. Maybe the p in 8p actually stands for perspective? You see the number 8 but I see an inverted duplicated letter p — we’re both right.
Biologists may benefit from borrowing a few quantum mechanics concepts from physicists. Chromosomes are objects in motion, after all. As we learn more about the dynamic 3-D architectures of genomes, the rules of chromosomal shape-shifting appear to be both deterministic and stochastic.
Chromosomes exhibit quantum-like duality of matter: compressing into arrays of compact condensates at one moment, and then, in the snap of a finger, splaying out into conformations choreographed by DNA replication and chromatin remodeling machinery that demands hitting temporal and spatial marks, i.e., specific chromosome-to-chromosome contacts.
A century after physics, biology needs the equivalent of a quantum description of the genome, where the software and hardware of life converge into a remarkable set of computational polymers called chromosomes. The static, staged 2-D rendering of karyotypes is a convenient simplification for diagnostic purposes but misses the 3-D chromosomal choreography that takes places during replication, transcription and DNA damage repair.
Chromosomes condense and decondense, reversibly and conditionally, in specific locations and times inside the nuclei of specific cells. What if any disturbance along the length of chromosome 8p alters the contacts made between chromosome 8 and itself as well as with other chromosomes, which would in turn perturb transcriptional programs that require those long-distance physical interactions? The technologies exist to map the 3-D genome architecture of chromosomes inside the nucleus and we plan to apply them thoughtfully this year.
The fact that 8p heroes nearly universally present with global neurodevelopmental delay tells us that neurogenesis is particularly vulnerable to traumatic genome injuries involving 8p. Something is happening to the genomes of stem cells in the brain as they contort into neurons.
Are the physical pressures of gene regulation forcing chromosomes together and the physical compacting forces on chromosomes inside the nucleus conspiring to create sensitivity to genomic imbalance in brain cells more so than other cells of the body throughout life? A 2023 paper from the Deisseroth lab at Stanford documented dramatic 3-D genome architectural changes in cerebellar granule cells, an understudied neuronal population that actually accounts for the vast majority of neurons in the human brain (Tan et al., 2023).
Of course we must also consider the effects of genomic imbalance on the rest of the cell outside of the nucleus. We know that increased demands on cellular metabolism, over-consumption of rate-limiting factors required for genome maintenance, and chronic activation of cellular stress responses ensue in the aftermath of genomic imbalance.
We’re not sure how all the puzzle pieces fit together. We’re certainly missing a few pieces right now that will surface as our data corpus grows, or that will become visible when we shift perspective. However, we think we’ve found the four corner pieces, meaning at this moment in the arc of 8p research we feel confident that the puzzle is solvable.
The p in 8p also stands for perseverance. We on the Science Team of Project 8p would like to think that we embody the spirit and the quest of the late Dr. Angelika Amon, who was a universally beloved scholar of aneuploidy.
Dr. Amon discovered the pillars of classical aneuploidy, which she viewed through the lens of cancer. Specifically, a dividing cell seeking a competitive growth advantage over its genetically identical neighbors, i.e., oncogenesis. Her work was so foundational and predictive that experiments performed in yeast cells (Saccharomyces cerevisiae) have been repeatedly corroborated in human cells and mouse embryos.
Had Dr. Amon not tragically passed away in 2020 her lab would surely be studying 8p as a means to deeper understanding of genomic imbalance, and just because she was such a kind-hearted person.
In fact, Dr. Amon received several 8p fibroblast lines from Dr. Chung’s lab. In what turned out to be a final parting gift, the Amon lab generated RNA-seq data comparing three invdupdel 8p fibroblast lines and three trisomy 21 fibroblast lines. These data are available on the online 8p data portal, which will ultimately be the home of all Project 8p-sponsored research.
In 2007, the Amon lab published a seminal paper describing a first-of-its-kind collection of yeast aneuploidy mutants harboring a gain or loss of each chromosome (Torres et al., 2007). On net, aneuploidy is a stressor. Aneuploid cells divide more slowly than euploid controls.
Even in such a simple system as yeast, three tenets of classical aneuploidy explain what is seen in the lab, and therefore yeast cells are predictive of the behavior of cancer cells in the human body:
An integrated stress response pathway that ramps up glycolysis, shuts down translation and increases protein degradation is chronically activated by imbalance of any chromosome (with a few exceptions based on the overall size of the chromosome).
Genes that change in DNA copy number (dosage) lead to proportional increases in mRNA levels which can be buffered at the protein level on a protein by protein basis but on average: more DNA leads to more mRNA which leads to more protein.
Specific dose-sensitive driver genes — sometimes just a single driver — are predominantly responsible for phenotypes rather than aneuploidy per se.
What constitutes a driver gene? Yeast and cancer cells behave similarly in “long term evolution” experiments, the former in the lab and the latter in the body. The mechanics of genome copying mean that even if one gene is solely responsible for a selective growth advantage, the cell can’t just selectively copy and paste parts of its genome on a strict gene-by-gene basis.
So, bystander genes down the street, chromosomally speaking, from a bona fide driver gene get carried along for the ride, say as part of a partial or whole-chromosome duplication event. The debate of what is a driver — and the extent to which aneuploidy itself is a driver of cellular or tissue pathology — has raged for decades in the oncology space.
Aneuploidy has been studied extensively in the context of cancer, which involves genome evolution of somatic cells. Genomic imbalance naturally occurs in individual cells of all humans as we age, usually taking decades to manifest as malignancy or decline in fitness.
Drs. Maitreya Dunham and David Botstein demonstrated in 2002 that yeast cells under selective pressure to grow faster experience the same kinds of chromosome copy number variations as precancerous cells in the human body (Dunham et al., 2002). In other words, aneuploidy occurs as a result of random errors in mitosis that increase in frequency as an organism ages.
But what happens when a traumatic genome injury occurs in meiosis, i.e., during the production of gametes (eggs or sperm), such that genomic imbalance is present in the first cell of a new life?
We all start out as a fertilized egg which multiplicatively blossoms into a fully formed fetus. In the case of 8p, the stresses of genomic imbalance and the shadowy actions of a conspiratorial network of drivers are experienced from the very first cell division, as one cell becomes two, two cells become four, and so on. Yet despite that congenital chromosomal wound, not all cells of the body are vulnerable to traumatic genome injuries involving 8p.
Dr. Amon’s lab showed that aneuploidy increases the rate of non-genetic individuality in cells, and this effect is amplified in whole organisms as they age (Beach et al., 2017). If you ask us, she was hot on the trail of a grand unified theory of chromosomal disorders. In our search for treatments and cures for 8p, we have taken up the banner in solidarity with a network of like-minded scientists in academia and industry.
In October 2022, Project 8p published the first edition of the 8p Cure Roadmap, a two-year research plan with four focus areas. Having passed the halfway point a few months ago, the primary purpose of this document is to highlight and synthesize key discoveries made by 8p-sponsored researchers. We also honor our open science commitment by sharing scientific and operational learnings that we hope will benefit other foundations focused on chromosome disorders.
Some scientific bets paid off, while others approaches will need to be revised or postponed or deprioritized. We share our strategy for expanding the 8p researcher network to include mission-aligned industry partners, e.g., biotech platform companies.
Building on the chromosome structure analogy, we originally described four research segments, to which we added a fifth segment: Assistive/Augmentative Devices.
Almost all 8p heroes have an AAC device in their daily lives. The idea of “electroceuticals” whose clinical development path is uncoupled from target-based therapeutic modalities is attractive from the point of view of delivering meaningful interventions to 8p heroes in the here and now.
We’re still in the exploration phase of getting to know academics and startups in the space. For example, we hosted a company called Actipulse at the October 2023 8p Research Roundtable. At the 8p Conference this summer, we plan on hosting a medical carnival where companies like Actipulse can demo their wares and interact with families and researchers at the same time.
Project 8p has provided catalytic funding to an academic researcher network comprised of five active projects (Meharena, Pinter, Sheltzer, Warmflash and Telese) and four completed projects (Chung, Muotri, Moisoi, and Logsdon/Eichler) as of February 2024.
The Project 8p Science Team is:
Ethan Perlstein, Ph.D. — Science Director
Barbara Celona, Ph.D. — Programs Director
Whitney Dolan, Ph.D. — Data Scientist
Segment 1: Drug Repurposing
Drug repurposing — finding a second or third (or nth) use for a drug already approved for one purpose — requires disease models that display pathological phenotypes that can be rescued in high-throughput assays.
We are primarily focusing on two complementary cell-based model systems: 8p skin fibroblasts, and 8p neural progenitor cells (NPCs) derived from reprogrammed iPSCs. We plan on deploying two unbiased assays: imaging of morphological features in cells by automated microscopy, and genome-wide measurements of mRNA expression levels in cells by automated sequencing.
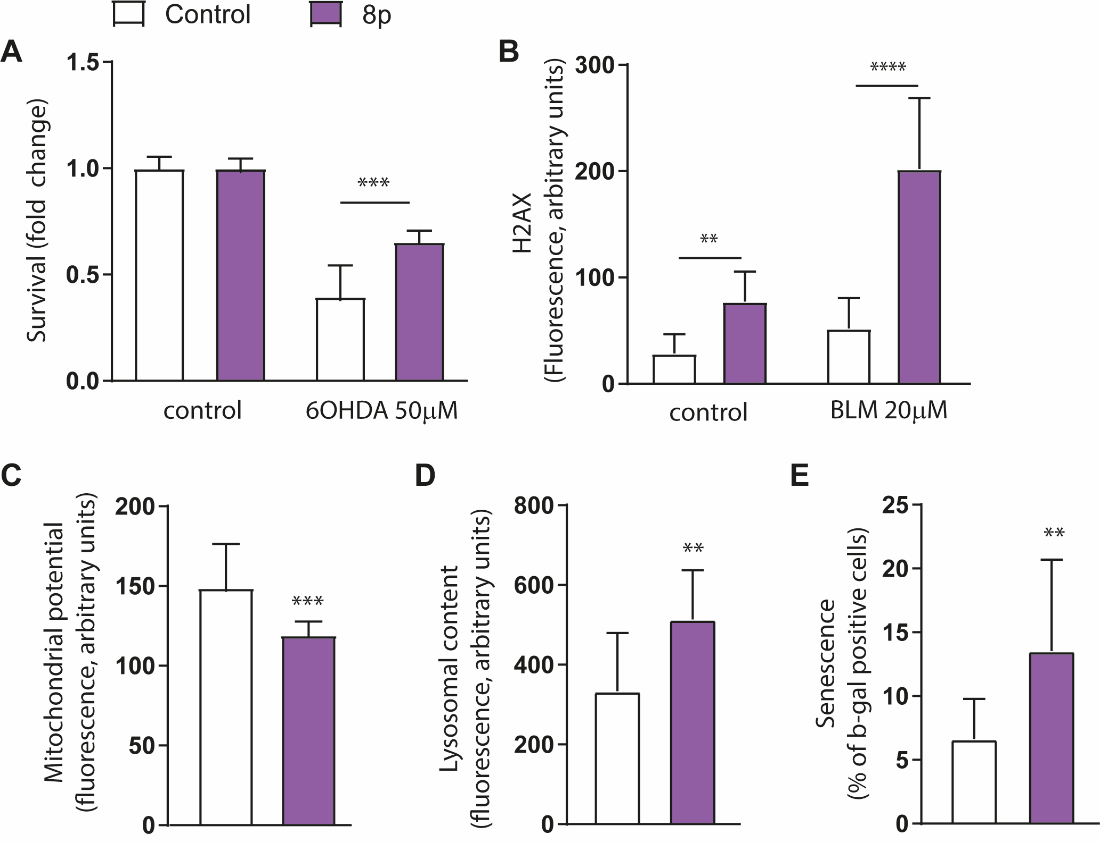
Project 8p worked with Dr. Marta Dominguez-Prieto in Dr. Nicoleta Moisoi’s lab in the UK to perform a rigorous battery of image-based and sequencing-based tests on a set of four 8p fibroblast lines — two invdupdels and two dels only — alongside four control fibroblast lines. The readouts included the Cell Painting palette, integrated stress response gene expression profiling, mitochondrial function assessments, DNA damage assessments, and senescence marker analysis.
Moisoi lab observed biological variability between the four 8p hero-derived fibroblast lines across all assays. In spite of that variability, statistically significant trends were detected when the four 8p samples are pooled and compared to the average of four healthy control lines.
For example, mitochondrial membrane potential is consistently decreased in the 8p samples. As another example, lysosomal content is consistently increased in the 8p samples. Intriguingly, the percentage of senescent cells is consistently increased in the 8p samples. (We’ll return the subject of senescence later).
Thanks to the proof-of-concept data generated by the Moisoi lab in a university setting, we are currently in discussions with a platform startup that has the ability to perform automated, AI-assisted, image-based drug screening on fibroblasts using Cell Painting and senescence markers.
The scaled up (and already paid for) capacity of a venture-backed startup compared to a single academic lab funded by a seed grant will enable us to test at least 10 different 8p fibroblast lines alongside a comparable number of controls. Ultimately, we may need to test dozens of different 8p fibroblast lines to overcome the biological variability intrinsic to 8p cells.
In parallel, we’re taking a second stab at drug repurposing using RNA-seq as the readout. Working with the company Rarebase taught us three complementary scientific lessons. First, a primary drug screen needed to be performed on 8p cells, not on healthy cells followed by hit validation in 8p cells. Second, the genome-wide transcriptional profile of 8p cells needed to be rescued, not just the expression of individual suspected driver genes. Third, pluripotent neural progenitor cells (NPCs) are a more therapeutically relevant substrate for high-throughput drug screening than terminally differentiated glutamatergic neurons.
Based on those learnings, we are currently in discussions with a platform startup to perform RNA-seq-based drug repurposing screens on 8p NPCs with the goal of restoring the genome-wide 8p transcriptional profile to normal, or as close to normal as possible.
We will elaborate further in the next section, but promising results from Dr. Alysson Muotri’s lab on cortical brain organoids open up the possibility of drug repurposing on a 3-D model, not just 2-D fibroblasts and NPCs. We’ve had preliminary discussions with a platform startup that has developed technologies for high-throughput screening to rescue germ layer patterning defects in 3-D brain organoids.
Segment 2: Disease Modeling
As described above in the tribute to Dr. Angelika Amon, the first forays into 8p disease modeling combined the simplicity and ruggedness of skin fibroblasts with RNA-seq. Three 8p invdupdel lines were compared to three trisomy 21 lines. We offered a high-level summary of those findings, juxtaposed with the Moisoi lab phenotypic assessments, last summer on our Substack (see Skin Deep).
At the other extreme of disease model complexity, Dr. Alysson Muotri’s lab cultured in the lab 1-month, 3-month, 6-month and 9-month cortical brain organoids from the pioneering family trio. Muotri lab then performed single-cell RNA-seq on all organoids from all timepoints, yielding a fecund dataset that we’re still exploring.
Although Muotri lab tested just a single clone of the 8p hero, maternal and paternal lines, we were delighted by an unexpected result: an unusual population of Cajal-Retzius-like neurons that express high levels of the gene Reelin (RELN) was present in the 8p hero organoids at the 6-month and 9-month timepoints. Strikingly, terminally differentiated excitatory glutamatergic neurons cultured for 21 days — but not for 7 days — express high levels of RELN, too. Those high-level findings were posted last year on our Substack in the practice of open science (see Reelin Groovy).
Open science isn’t just a buzzword, it’s a proven research accelerant and an effective researcher recruitment tool. We would not have appreciated the Reelin convergence between 3-D cortical brain organoids and 2-D glutamatergic neurons had it not been for Project 8p’s monthly 8p Research Roundtables, where those data were shared hot off the presses.
Dr. Francesca Telese read the Reelin Groovy post and was inspired to contact us. That spark led to our current exciting bioinformatics collaboration with Dr. Yanning Zuo in the Telese lab. The goal of this project is to extract biological insights from the massive cortical organoid single-cell RNA-seq experiment completed by the Muotri lab. Where those insights will take us beyond Reelin and Cajal-Retzius neurons is anyone’s guess at this point, but that’s the nature of research on the frontier of knowledge.
We now have RNA-seq datasets from 8p fibroblasts, 8p neural progenitor cells, 8p differentiated excitatory neurons, and 8p cortical brain organoids for a pioneering invdupdel 8p hero. With our expanded Science Team in place and the launch of the 8p data portal in the weeks ahead, we will perform a comprehensive analysis and immediately share the results with our 8p researcher network.
Dr. Hiruy Meharena’s lab was recruited by Project 8p based on his wealth of experience with trisomy 21 and his integrative approach to studying chromosomal disorders that share a core set of clinical features.
So far, Co-Investigator Dr. Ashley Watson and teammates in the Meharena Lab have transcriptionally profiled 8p neural progenitor cells (NPCs) from the pioneer invdupdel proband side-by-side with trisomy 21 neural progenitor cells.
Incredibly, half of differentially expressed genes in 8p NPCs are identical in trisomy 21 NPCs! Cooler still, we observe the same degree of transcriptional convergence between 8p fibroblast lines and trisomy 21 fibroblast lines.
Rewinding the developmental clock, Project 8p sought out Dr. Aryeh Warmflash to study the first stages of brain development using induced pluripotent stem cells differentiated toward ectoderm, the embryonic tissue that in part gives rise to the brain.
The Warmflash lab collected data on the pioneer 8p invdupdel line showing a clear phenotype of increased mesoderm differentiation. His lab honed in on two 8p genes, FGF17 and TNKS, and assayed their effect on germ layer differentiation. The final results of this project will be published on our Substack soon.
What’s next on the disease modeling menu? We’ve got flies, zebrafish, frogs, and mice. All of those animal models have demonstrated their chops in monogenic disease modeling, but the lack of synteny across evolutionary timescales means human chromosome 8 is not the same as mouse chromosome 8, and forget about comparisons between human and the simpler model organisms. We’re in active discussions with academic labs and platform startups to determine the smartest proof-of-concept projects.
Segment 3: Chromosome Therapy
Acknowledging that chromosome therapies are on the distant horizon due to delivery challenges and the need for persistence over cell division, we are focusing on the practical: a toolkit for chromosome engineering.
The most important tool is a so-called isogenic control. In the case of a monogenic variant, CRISPR can be used to correct the disease-causing mutation while in theory not leaving a meaningful scratch anywhere else in the genome. But there is no gene editing machinery that can operate on the scale of megabases. The state of the art today, prime editing maxes out in the tens of kilobases. Instead we must rely on laboratory evolution.
Project 8p enlisted Dr. Jason Sheltzer to the 8p cause with the goal of isolating and characterizing an isogenic control of the pioneering invdupdel 8p hero induced pluripotent stem cell (iPSC) line. Dr. Sheltzer trained with Dr. Amon and his day job is the study of aneuploidy in the context of cancer.
Sophia Lee in the Sheltzer lab tried multiple approaches to restore disomy to the 8p iPSC line. We were looking for a needle in the haystack: a single 8p cell in which the rearranged chromosome 8 is lost and replaced by a second copy of the un-rearranged chromosome.
What seemed to work was the genomic equivalent of a Texas two-step. The un-rearranged copy of chromosome 8 got duplicated resulting in an unstable interim trisomic state that resolved back to disomy by subsequent shedding of the rearranged copy of chromosome 8. These results are published on our Substack (see Balancing Act).
Technically, the new ground state is called uniparental disomy, the closest we’ll get to an isogenic control short of performing reconstructive molecular surgery on the actual rearranged copy of chromosome 8. We’ll know if a presumptive isogenic corrected clone is free of any unwanted genomic alterations soon.
This point can’t be stressed enough: having an 8p isogenic control means in future experiments we’ll have a near-perfect comparison to the original 8p line, and won’t have to rely either on genetically unrelated age-matched and sex-matched controls or on parental controls. Eventually, Project 8p will support the generation of isogenic controls for all 8p hero iPSC lines.
To round out the chromosome engineering toolkit, we recruited Dr. Stefan Pinter to 8p based on his unique thoughtfulness and experience with another chromosomal disorder called Turner Syndrome. The Pinter lab has engineered a euploid (healthy) iPSC line and an 8p invdupdel iPSC line to express either a CRISPR interference (CRISPRi) construct enabling specific gene knockdown, or a CRISPR activation (CRISPRa) construct enabling specific gene activation.
The CRISPRi lines are ready for functional screens. The CRISPRa lines need further optimization but will hopefully be ready to deploy soon. An NGN2 cassette was added to said CRISPRi/a lines to permit differentiation of iPSCs into excitatory neurons. A proposal for an RNA-seq-based functional genomics screen that interrogates multiple candidate 8p drivers using the CRISPRi and CRISPRa lines is currently being evaluated. In parallel, plans are afoot to use XIST-mini genes to silence 1-2 megabase stretches at a time.
Many others are racing to solve the artificial chromosome synthesis problem and the artificial chromosome in vivo delivery problem. Where Project 8p could make a difference is by supporting proof-of-concept megabase-plus chromosome engineering in mouse stem cells and the eventual generation of the first bona fide 8p mouse model.
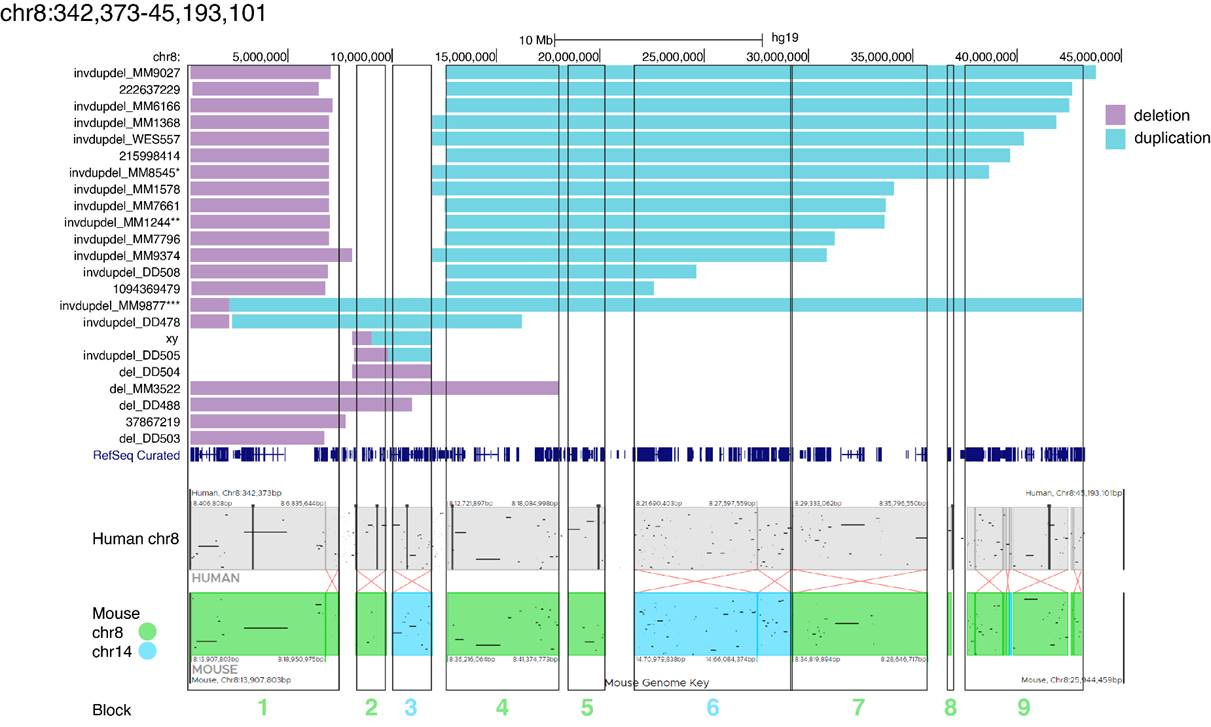
We think an ideal academic partner in this endeavor would be Dr. Jef Boeke’s lab at NYU based on their extensive success at chromosome engineering in yeast cells. Dr. Ran Brosh from the Dark Matter Project generated the figure below that shows conserved synteny blocks between human chromosome 8 and mouse chromosome 8 along the length of 8p.
A “Block 1” mouse may — we emphasize may — model 8p heroes with a distal deletion only. Even if we successfully obtain the deletion and observe a phenotype, interpreting and understanding whether it relates to 8p heroes will be challenging —and mice are expensive to maintain. We’re currently in early discussions to refine the scope of work and discharge as much risk upfront as possible.
We would also like to collaborate with Dr. Glennis Logsdon, now at University of Pennsylvania, who was funded by Project 8p during her time in Dr. Evan Eichler’s lab. She was eager to explore novel approaches on chromosome engineering directly in 8p cells.
Segment 4: In hero studies
An IRB approved longitudinal natural history study and biorepository has been expanding beyond the Chung study under the oversight of founder, Bina Maniar Shah, in dutiful service to the community as Principal Investigator. Now over 150 heroes are enrolled and caregivers with curated genetic breakpoints linked to clinical phenotypic data.
Much of the behind-the-scenes, unglamorous and often tedious work is attributed to Patient Engagement Manager, Kaiti Syverson, mom to 8p hero Chloe, and Dr. Whitney Dolan, staff data scientist. The diverse biosample collection is a powerful research asset with multiple clones and gender matched biological parent or sibling that will bring new stakeholders into the fold.
We are putting the finishing touches on the 8p Insights Portal where researchers will be able to compare de-identified genotype and phenotype data to form cohorts and hypotheses, as well as to request biosamples and experimental data to put their hypotheses to the test. This endeavor was partly inspired by the portals constructed by Dr. Dennis Lal’s lab for neurodevelopmental disorders, both monogenic and chromosomal. With every 8p hero affected differently and layering publicly available human phenotype ontology and eventually ML algos, you can see the endless possibilities of sleuthing for therapeutic targets.
In a collaboration with Illumina, an exciting data set and analysis is evolving and being made available to researchers. This genotype-phenotype set includes clinician reported data with whole genomes, RNA seq, transcriptomes, and DNA methylomes from peripheral blood mononuclear cells from 100 families across 8p, Dup15q, and Ring14 that will be interpreted and compared.
And none of this would be possible without our research partners’ commitment to open science making discovery data available.
In the concluding section below, we’ll share a synopsis of the first ever “in hero” interventional nutraceutical study that will be sponsored and managed by Project 8p.
Duplicated 8p drivers of interest
We end this roadmap refresh by going a bit deeper on a few scientific concepts that we broached above, starting with driver genes that call 8p home. What follows is hypothesis and speculation, and a sprinkle of thought provocation.
Sajan et al., 2013 and Vibert et al., 2021 identified two overlapping duplicated 8p segments linked to anomalies or agenesis of the corpus callosum, which is observed at a frequency of 63% in a cohort of 27 8p heroes published just after Okur et al. A 5-megabase critical region includes 51 coding genes. One can make an argument for several of them.
First and foremost is the gene RHOBTB2. RHOBTB2 missense mutations are associated with a neurodevelopmental disorder similar in clinical presentation to 8p. Next on the list are the genes NEFL and NEFM, which encode neurofilament light chain and neurofilament medium chain, respectively. Mutations in NEFL and NEFM cause Charcot-Marie Tooth disease, which is a neurodegenerative condition. As a final example, the gene EBF2. EBF2 in mice controls the migration of Purkinje cells in the cerebellum, and GnRH neurons from the olfactory epithelium to the hypothalamus. Tying back to the RELN finding from the cortical brain organoids, EBF2 is also needed for the generation and migration of Cajal-Retzius cells.
We will use NGN2-induced 8p neurons with inducible CRISPRi to examine cellular and neuronal phenotypes of overexpression of candidate triplo-sensitive 8p driver genes like RHOBTB2, NEFL, NEFM and EBF2.
Genomic imbalance leads to genome dyshomeostasis
The prevailing wisdom about genomic imbalance is that out of the three — DNA, RNA, protein — the real culprit driving aneuploidy stress are out-of-whack protein levels. But what about the total levels of DNA? RNA?
The graduate work of Dr. Barbara Celona on our Science Team, where she studied chromatin remodeling and genome homeostasis, is coincidentally informative to our thinking about chromosomal disorders. We know from classical experiments that aneuploidy per se, i.e., extra and unplanned genomic DNA, is a universal cellular stress, or more accurately a universal collection of linked stresses.
All of that extra and unplanned DNA needs to be wrapped up into nucleosomes by proteins called histones. But it’s not as though the cell has an infinite supply of histones or the wherewithal to manufacture more histones on-demand without incurring unacceptable metabolic or biosynthetic tradeoffs. Also, a damaging loop ensues where DNA not bound by histones is more susceptible to damage and more likely to be inappropriately transcribed.
We’re just starting to wrap our minds around which features of 8p pathology are caused by discrete gene drivers and which features are caused by the cascading effects of genomic imbalance on cell health and survival. The truth is likely a complex interplay of both sets of features.
All segments point to neurogenesis
While the heart and skeletal systems are affected in 8p and other chromosomal disorders, the most affected organ is the brain, especially during fetal and early childhood neurodevelopmental. Why are neural progenitor cells (NPCs) so vulnerable to 8p genomic imbalance and more generally?
We can only make conjectures at this juncture. One consideration surely is the hard-wired metabolic shift from glycolysis to mitochondrial-dependent oxidative phosphorylation (OxPhos) that occurs in NPCs as they differentiate into committed neuronal or glial cell lineages.
Genomic imbalance is a metabolic imposition, an unplanned consumption of rate-limiting metabolites. Neurogenesis may be unique among all the 200+ cell fates possible in the human body because of the rarified combination of morphological and functional adaptations that only neurons seemingly possess. One of those is hypersensitivity to disruptions in metabolic supply chains.
From neurodev to neuropsych
8p heroes with distal dups tend be to mild-to-moderate in terms of clinical presentation. Their neurological symptoms manifest more as autism-spectrum disorder and attention deficit disorder. Project 8p will begin this year to broaden our scope beyond early childhood neurodevelopment.
Based on hints and clues from other chromosomal disorders, we cannot ignore that as 8p heroes age there needs to be in place proactive surveillance for neuropsychiatric and possibly neurodegenerative symptoms. Over the next few months will we will have conversations with appropriate experts in the field.
Integrated Stress Response and Senescence
A landmark 2019 paper from the labs of Drs. Peter Walter and Mauro Costa-Mattioli implicated the integrated stress response (ISR) pathway is a driver of disease pathology in trisomy 21 aka Down Syndrome (Zhu et al., 2019). Combined with the knowledge of Dr. Meharena’s work on trisomy 21 NPCs, senescence was on our radar, too.
The Moisoi lab results consisting of four 8p fibroblast lines indicated that, on the mRNA level at least, levels of canonical ISR pathway markers are modestly decreased.

We are in contract discussions with a platform startup to discover novel small molecule modulators of ISR and senescence, which could serve as both 8p medicines and one day in the future anti-aging drugs.
Minimum Viable Clinical Trial
Collaborating with nutritionist Geri Brewster RD MPH CDN, we are putting in place the key ingredients of a patient-driven nutraceuticals trial that will establish a robust regiment to optimize development with over-the-counter supplements for all 8p heroes. That way all 8p heroes start out on a more level playing field for foundational health and nutrients. At the same time, we maximize benefit from interventions that don’t require FDA approval. Protocol drafting will commence next month.
Project 8p will act as the trial sponsor with respect to IRB approval. We anticipate recruiting a dozen 8p heroes to participate. We are in discussions with health tech companies and developers for an app or browser-based tool to help us collect trial data easily with a reduced burden on caregivers. It can include electronic health records and using simple mobile phone based video and survey assessments. This in itself will help us go deeper with some families on our natural history study but also invent tools for clinical outcome measures and endpoint analysis.
We’re not holding our breath that a nutraceutical cocktail will be a magic elixir. This exercise is about building the muscle memory and lived experience of 8p heroes and their families so that when a drug repurposing or novel drug candidate or an assistive device/drug combination therapy is ready for prime time, so is the 8p community. Let’s do this!
[The next edition of the 8p roadmap will be a team science co-authored effort written by the end of 2024 and take us through the end of the decade. It will serve as the “roadmap of record” in a peer-reviewed journal.]